Abstract
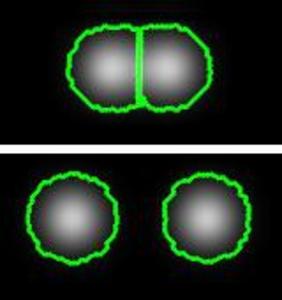
An Insight Toolkit (ITK) processing framework for segmenting and tracking nuclei in time-lapse microscopy images using coupled active contours is presented in this paper. We implement the method of Dufour et al.[2] to segment and track cells in fluorescence microscopy images. The basic idea is to model the image as a constant intensity background with constant intensity foreground components. We utilizes our earlier submissions on the Chan and Vese algorithm [1] and its multiphase extension [5] to build our new tracking filter. The tracking filter itk::MultiphaseLevelSetTracking inputs a segmentation result (or a coarse estimate) from the previous time-point along with the feature image and generates a new segmentation output. By iteratively repeating this process across all time-points, real-time tracking is made possible. We include 2D/3D example code, parameter settings and show the results generated on a 2D zebrafish embryo image series.
Keywords
Source Code and Data
Loading file tree...